First Variant was the D614G Mutation
While the D614G mutation was relatively rare at the onset of the pandemic, by June 2020 this mutation was found in the majority of COVID-19 patients. The mutation consisted of an amino acid change from aspartate to a glycine residue at position 614 (D614G). see: Biosensing Detection of the SARS-CoV-2 D614G Mutation
The D614G mutation is located in the S1 subunit of the S protein (amino acids 14–685), which is crucial for the entry of the SARS-CoV-2 virus into the host cell (Figure 1 below). The S protein of SARS-CoV-2 is cleaved into S1 and S2 subunits by host proteases, with the S1 domain being responsible for receptor binding and the S2 domain for mediating membrane fusion -- The receptor binding domain (RBD) within the S1 subunit of S protein interacts with the angiotensin-converting enzyme 2 (ACE2) receptor on the surface of the host cells, thus mediating virus entry. Although, the D614G point mutation is outside of the RBD (Figure 1), it has been suggested to have an impact on the infectivity of the virus see: (Zhang, 2020 and references therein).
The rapid spread of the D614G mutant version of the virus was hypothesized to correlate with the higher infectivity of SARS-CoV-2 harboring this mutation. Clinical samples from patients infected with the D614G mutant were shown to have higher viral RNA levels and higher pseudovirus titers... Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus
Figure 1.
The location of amino acid 646 is in the S1 region of the spike protein--(amino acids 14–685)
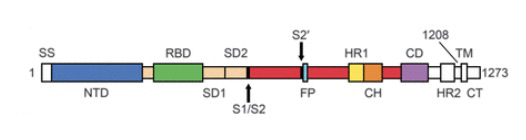
Above image: shows amino acids labeled showing NTD (N-Terminal Domain), RBD (Reaction Binding Domain) located in S1 domain and components of S2 domain. For a full explanation see: Wrapp et al., (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation.
What are the Effects of D614G at Molecular Level?
Cryo EM studies --Gobeil et al., (2021) D614G Mutation Alters SARS-CoV-2 Spike Conformation and Enhances Protease Cleavage at the S1/S2 Junction --
"...Cryoelectron microscopy (cryo-EM) structures reveal altered receptor binding domain (RBD) disposition; antigenicity and proteolysis experiments reveal structural changes and enhanced furin cleavage efficiency of the G614 variant. Furthermore, furin cleavage alters the up/down ratio of the RBDs in the G614 S ectodomain, demonstrating an allosteric effect on RBD positioning triggered by changes in the SD2 region, which harbors residue 614 and the furin cleavage site...."
--D614G mutation increases RBD "up" state and enhances S1/S2 junction proteolysis
--D614G— seems to relax connections between peptides. This makes open conformations (RBD up) more likely and might increase the chance of infection
-- Negative stain electron microscopy revealed a higher percentage of the 1-RBD "up" conformation in the G614 spike, suggesting increased epitope exposure as a mechanism of enhanced vulnerability to neutralization..." D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization
The SARS-CoV-2 Spike Variant D614G Favors an Open Conformational State
"...The COVID-19 pandemic underwent a rapid transition with the emergence of a SARS-CoV-2 variant that carried the amino acid substitution D614G in the Spike protein that became globally prevalent. The G-form is both more infectious in vitro and associated with increased viral loads in infected people. To gain insight into the mechanism underlying these distinctive characteristics, we employed multiple replicas of microsecond all-atom simulations to probe the molecular-level impact of this substitution on Spike’s closed and open states. The open state enables Spike interactions with its human cellular receptor, ACE2. Here we show that changes in the inter-protomer energetics due to the D614G substitution favor a higher population of infection-capable (open) states. The inter-protomer interactions between S1 and S2 subunits in the open state of the D-form are asymmetric. This asymmetry is resolved in the G-form due to the release of tensile hydrogen bonds resulting in an increased population of open conformations. Thus, the increased infectivity of the G-form is likely due to a higher rate of profitable binding encounters with the host receptor. It is also predicted to be more neutralization sensitive due to enhanced exposure of the receptor binding domain, a key target region for neutralizing antibodies..." source --Mansbach RA et al.
Molecular Modeling of the D614G region
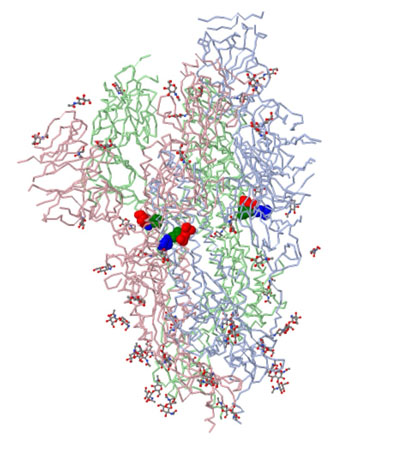
Spike protein in prefusion conformation with single rbd up
PDB
FILE: 6VSB
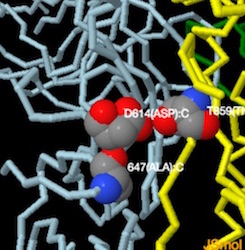
Image above shows the site of the first D614G mutation ---amino acids of interest in CPK: Asp614 (blue), Thr859 (green) and Lys854 (red).
Cai et al., ---Distinct conformational states of SARS-CoV-2 spike protein
The atomic structure coordinates are deposited in the RCSB Protein Data Bank (PDB) under the accession numbers 6XR8 -- has salt bridge binding d614 to lysine 854 ----and 6XRA --(post fusion structure); and the electron microscopy maps have been deposited in the Electron Microscopy Data Bank (EMDB)
Structural transition from the prefusion to postfusion conformation inducing membrane fusion likely proceeds stepwise as follows. First, FPPR clamps down RBD through CTD1 in the prefusion S trimer (this study) but occasionally flips out of position and allows an RBD to sample the up conformation (PDB ID: 6vyb --there are a lot of disordered amino acids around d614) in PDB structures.
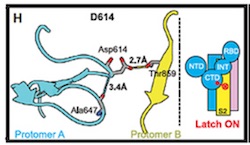
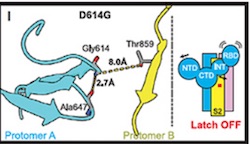
Based on the models derived from cryo-EM, it was hypothesized that the mutation results in the loss of a hydrogen bond interaction between Asp614 and Thr859, which resides in different S monomers.see: source.
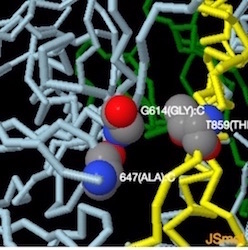
"...Substitution of Asp614 with glycine changes hydrogen bonding around residue 614. In the case of D614 (L), an inter-protomer hydrogen bond is detected. For D614G (M), the Asp614-Thr859 hydrogen bond is eliminated, and interaction with intradomain Ala647 is strengthened...." Yurkovetskiy et al.,
Image below from Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant--Yurkovetsky et al., shows Asp614 hydrogen bonded to Thr859.(Image H andI) Substitution of Asp614 with glycine changes hydrogen bonding around residue 614. In the case of D614 (H), an inter-protomer hydrogen bond is detected. For D614G (I), the Asp614-Thr859 hydrogen bond is eliminated, and interaction with intradomain Ala647 is strengthened. See also Figures S1 and S2 in source.
PDB: 6VSB D614
-------------->spin on -------->- spin off
->zoom in to D614:C -------->- zoom out
To Rotate the Molecule--->Left Click and Drag
To Zoom-->>Left Click + hold Shift button and Drag Vertically
To see amino acid and atom number hold cursor over atom
Jmol Menu --->>Right-Click
PDB:7KDL-->D614G
-------------->spin on -------->- spin off
->zoom in to D614:G -------->- zoom out
To Rotate the Molecule--->Left Click and Drag
To Zoom-->>Left Click + hold Shift button and Drag Vertically
To see amino acid and atom number hold cursor over atom
Jmol Menu --->>Right-Click
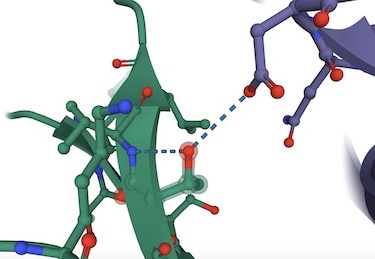
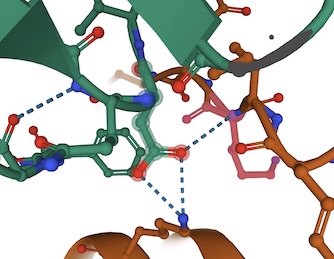
Left: pdb 6VSB--Wrapp et al., Prefusion 2019-nCoV spike glycoprotein with a single receptor-binding domain up (Aspartic 614 H-bonded to Threonine 859 and Alanine 647)-- . Right: pdb 6XR8-- Cai et al., Distinct conformational states of SARS-CoV-2 spike protein --- lysine 854 (B Chain) in a salt bridge with Aspartic acid 614(A Chain) Note: 7DF3-pdb (xu et al., 2021)also has salt bridge,
The weakening of the G614 bonding (hydrogen bonds and/or salt bridge) shown above results in increased infectivity of the virus.
Despite the increased infectivity of D614G in tissue culture, and the increased viral load in infected people, increased COVID-19 disease severity has not been detected in association with D614G infection
Sars-CoV-2 Variants
The novel coronavirus SARS-CoV2
B.1.1.7 and B.1.525 UK variant
B.1.427/B.1.429, California QP77P Mutation
B.1.526, NY E484K or S477N Mutations
Molecules of Disease
-
Small Molecules
Cholesterol
Nicotine
Trans Fatty Acid
Alcohol
Acetyaldehyde
- Proteins
Cytokines
Hemoglobin S
Prions
Botulinum Toxin
Explain it with Molecules
- Why is water such a good solvent?
- Why does ice float?
- Why do solids, liquids and gases behave differently?
- What is the geometry of methane?
- What's the difference between alpha and beta glucose?
- How does caffeine work in the brain?
- How does soap work?
- What is the difference between sucrose and fructose?
- Why is carbon monoxide so dangerous?
- Why is graphite so soft if it is made of only carbon?
- What is the difference between Carbyne and Graphite?
- Why is the fullerene and similar structures the cornerstone of nanotechnology?
- How big is a nanotube?
- Why does table salt have a cubic crystal shape?
- What is the structure of the benzene molecule?
- Why do carcinogens cause cancer?
- What causes Sickle Cell Anemia?
- What is the difference between sodium nitrite and nitrate?
- How do drugs work?