UK Variants-B.1.1.7 and B.1.525
B.1.1.7 lineage (a.k.a. 20I/501Y.V1 Variant of Concern (VOC) 202012/01) - source CDC.gov
- This variant has a mutation in the receptor binding domain (RBD) of the spike protein at position 501, where the amino acid asparagine (N) has been replaced with tyrosine (Y). The shorthand for this mutation is N501Y. This variant also has several other mutations, including:
- 69/70 deletion: occurred spontaneously many times and likely leads to a conformational change in the spike protein
- P681H: near the S1/S2 furin cleavage site, a site with high variability in coronaviruses. This mutation has also emerged spontaneously multiple times.
- This variant is estimated to have first emerged in the UK during September 2020.
- Since December 20, 2020, several countries have reported cases of the B.1.1.7 lineage, including the United States.
- This variant is associated with increased transmissibility (i.e., more efficient and rapid transmission).
- In January 2021, scientists from UK reported evidence[1] that suggests the B.1.1.7 variant may be associated with an increased risk of death compared with other variants.
- Early reports found no evidence to suggest that the variant has any impact on the severity of disease or vaccine efficacy. source
N501Y, enhances spike binding at the human ACE2 cell receptor
"...Using all atom molecular dynamics simulations, we demonstrated that Y501 in mutated RBD can be well coordinated by Y41 and K353 in hACE2 through hydrophobic interactions, increasing the overall binding affinity between RBD and hACE2 by about 0.81 kcal/mol..." Luan et al.,
Molecular Modeling of Y501 to ACE2 receptor
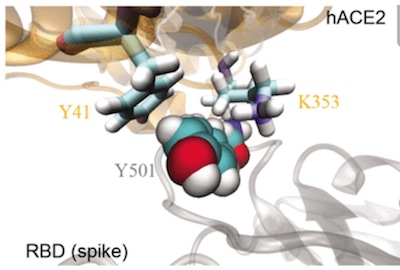
Above from Luan et al. shows increased binding with Y501 mutation to ACE2 receptor
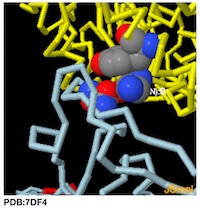
PDB:7DF4
-------------->spin on -------->- spin off
->zoom in to N501:B -------->- zoom out
To Rotate the Molecule--->Left Click and Drag
To Zoom-->>Left Click + hold Shift button and Drag Vertically
To see amino acid and atom number hold cursor over atom
Jmol Menu --->>Right-Click
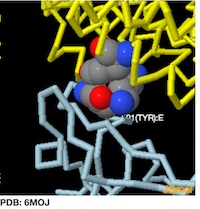
PDB: 6MOJ
-------------->spin on -------->- spin off
->zoom in to Y501:E -------->- zoom out
To Rotate the Molecule--->Left Click and Drag
To Zoom-->>Left Click + hold Shift button and Drag Vertically
To see amino acid and atom number hold cursor over atom
Jmol Menu --->>Right-Click
Sars-CoV-2 Variants
The novel coronavirus SARS-CoV2
B.1.1.7 and B.1.525 UK variant
B.1.427/B.1.429, California QP77P Mutation
B.1.526, NY E484K or S477N Mutations
Molecules of Disease
-
Small Molecules
Cholesterol
Nicotine
Trans Fatty Acid
Alcohol
Acetyaldehyde
- Proteins
Cytokines
Hemoglobin S
Prions
Botulinum Toxin
Explain it with Molecules
- Why is water such a good solvent?
- Why does ice float?
- Why do solids, liquids and gases behave differently?
- What is the geometry of methane?
- What's the difference between alpha and beta glucose?
- How does caffeine work in the brain?
- How does soap work?
- What is the difference between sucrose and fructose?
- Why is carbon monoxide so dangerous?
- Why is graphite so soft if it is made of only carbon?
- What is the difference between Carbyne and Graphite?
- Why is the fullerene and similar structures the cornerstone of nanotechnology?
- How big is a nanotube?
- Why does table salt have a cubic crystal shape?
- What is the structure of the benzene molecule?
- Why do carcinogens cause cancer?
- What causes Sickle Cell Anemia?
- What is the difference between sodium nitrite and nitrate?
- How do drugs work?