Delta Variant B.1.617.2
SARS-CoV-2 Delta variant has rapidly replaced the Alpha variant (also known as lineage B.1.1.7) around the world. The Delta variant B.1.617.2 is a sublineage of the Pango B.1.617 linage which also contains the SARS-CoV-2 Kappa variant which is also known as lineage B.1.617.1. The Kappa variant was first detected in India in December 2020. By the end of March 2021, the Kappa sub-variant accounted for more than half of the sequences being submitted from India. The Kappa variant has three notable alterations in the amino-acid sequences, all of which are in the virus's spike protein code namely: L452R, E484Q, P681R.
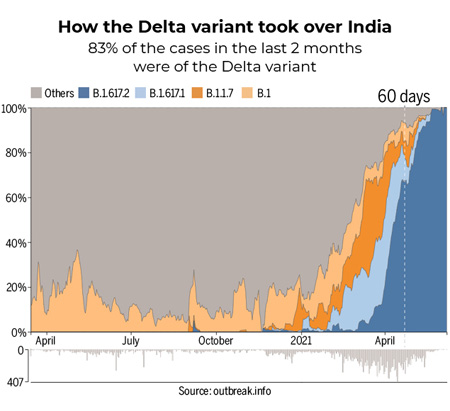
As shown in the chart below the Delta variant took over rapidly in India starting April 2021 and spread rapidly throughout the world. It is now the predominant strain in the US.
The Delta/ B.1.617.2 genome has 15-17 mutations depending on whether more common mutations are included which produce alterations in the amino-acid sequences of the proteins it encodes.
Four of them, all of which are in the virus's spike protein code, are of particular concern:
D614G. The substitution at position 614, an aspartic acid-to-glycine substitution, is shared with other highly transmissible variants like Alpha, Beta and Gamma.
T478K. The exchange at position 478 is a threonine-to-lysine substitution.
L452R. The substitution at position 452, a leucine-to-arginine substitution, confers stronger affinity of the spike protein for the ACE2 receptor[38] and decreased recognition capability of the immune system.
P681R. The substitution at position 681, a proline-to-arginine substitution, which, according to William A. Haseltine, may boost cell-level infectivity of the variant "by facilitating cleavage of the S precursor protein to the active S1/S2 configuration". Note: B.1.1.7 has P681H: near the S1/S2 furin cleavage site, a site with high variability in coronaviruses. This mutation has also emerged spontaneously multiple times.
The E484Q mutation is not present in the B.1.617.2 genome but is in B.1.617.1
How does the virus enter the cell?
"...SARS-CoV-2 S protein binds to the ACE2 receptor on the host cell, initially through the S1 receptor binding domain. The S1 domain is then shed from the viral surface, allowing the S2 domain to fuse to the host cell membrane. This process is dependent upon activation of the S protein, by cleavage at two sites (S1/S2 and S2’) via the proteases Furin and TMPRSS2. Furin cleavage at the S1/S2 site may lead to conformational changes in the viral S protein that exposes the RBD and/or the S2 domain. TMPRSS2 cleavage of the SARS-CoV-2 S protein is believed to enable the fusion of the viral capsid with the host cell to permit viral entry..." source --Structural and functional mechanism of SARS-CoV-2 cell entry
Note: The S1 Subunit Is Heavily Involved in Host Cell Receptor Binding while The S2 Subunit Is Heavily Involved in Membrane Fusion
Importance of Furin cleavage site
...Delta SARS-CoV-2 bearing the Alpha-spike glycoprotein replicated less efficiently than the wild-type Delta variant, suggesting the importance of Delta spike in enhancing viral replication. The Delta spike has accumulated mutation P681R located at a furin cleavage site that separates the spike 1 (S1) and S2 subunits. Reverting the P681R mutation to wild-type P681 significantly reduced the replication of Delta variant, to a level lower than the Alpha variant. Mechanistically, the Delta P681R mutation enhanced the cleavage of the full-length spike to S1 and S2, leading to increased infection via cell surface entry. In contrast, the Alpha spike also has a mutation at the same amino acid (P681H), but the spike cleavage from purified Alpha virions was reduced compared to the Delta spike. Collectively, our results indicate P681R as a key mutation in enhancing Delta variant replication via increased S1/S2 cleavage. Spike mutations that potentially affect furin cleavage efficiency must be closely monitored for future variant surveillance. from Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant
.. In fact, the S-protein of SARS-CoV-2 has a furin cleavage site that is missing in SARS-CoV. This is a section in the protein with the amino acid sequence proline-arginine-arginine-alanine, which is referred to as the PRRA motif or as polybasic cleavage site (PCS) (also referred to as furin cleavage site). The gain of this sequence with the neighboring furin cleavage site is likely responsible for the high infectivity of SARS-CoV-2, because the binding strength to the ACE2 receptor is similar to SARS-CoV, which is less infectious.
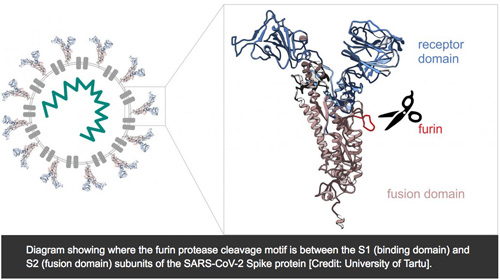
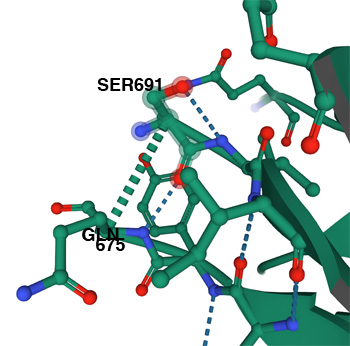
Image below: does not show critical amno acid 681R. The region GLN675- Ser691 is likely cleaved or disordered.
"...Pre-activated virus To penetrate cells, the SARS-CoV-2 spike protein must be cut twice by host proteins. In the SARS-CoV-1 virus that causes severe acute respiratory syndrome (SARS), both incisions occur after the virus has locked on to a cell. But with SARS-CoV-2, the presence of the furin cleavage site means that host enzymes (including one called furin) can make the first cut as newly formed viral particles emerge from an infected cell. These pre-activated viral particles can then go on to infect cells more efficiently than do particles requiring two cuts, says Whittaker...."
"...Shi’s team and other groups have zeroed in on a mutation that alters a single amino acid in the SARS-CoV-2 spike protein — the viral molecule responsible for recognizing and invading cells. The change, which is called P681R and transforms a proline residue into an arginine, falls within an intensely studied region of the spike protein called the furin cleavage site..."
Follow-up experiments by both groups showed that the P681R change was largely responsible for spike being clipped so much more efficiently.
Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant
"...The Delta spike has accumulated mutation P681R located at a furin cleavage site that separates the spike 1 (S1) and S2 subunits. Reverting the P681R mutation to wild-type P681 significantly reduced the replication of Delta variant, to a level lower than the Alpha variant. Mechanistically, the Delta P681R mutation enhanced the cleavage of the full-length spike to S1 and S2, leading to increased infection via cell surface entry. .."
How the coronavirus infects cells — and why Delta is so dangerous
SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India
Abstract: As the global severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic expands, genomic epidemiology and whole genome sequencing are being used to investigate its transmission and evolution. Against the backdrop of the global emergence of “variants of concern” (VOCs) during December 2020 and an upsurge in a state in the western part of India since January 2021, whole genome sequencing and analysis of spike protein mutations using sequence and structural approaches were undertaken to identify possible new variants and gauge the fitness of the current circulating strains. Phylogenetic analysis revealed that newly identified lineages B.1.617.1 and B.1.617.2 were predominantly circulating. The signature mutations possessed by these strains were L452R, T478K, E484Q, D614G and P681R(Note: pdb:7v7f file shown here has 478 with Threonine) in the spike protein, including within the receptor-binding domain (RBD). Of these, the mutations at residue positions 452, 484 and 681 have been reported in other globally circulating lineages. The structural analysis of RBD mutations L452R, T478K and E484Q revealed that these may possibly result in increased ACE2 binding while P681R in the furin cleavage site could increase the rate of S1-S2 cleavage, resulting in better transmissibility. The two RBD mutations, L452R and E484Q, indicated decreased binding to select monoclonal antibodies (mAbs) and may affect their neutralization potential. Further in vitro/in vivo studies would help confirm the phenotypic changes of the mutant strains. Overall, the study revealed that the newly emerged variants were responsible for the second wave of COVID-19 in Maharashtra. Lineage B.1.617.2 has been designated as a VOC delta and B.1.617.1 as a variant of interest kappa, and they are being widely reported in the rest of the country as well as globally. Continuous monitoring of these and emerging variants in India is essential.
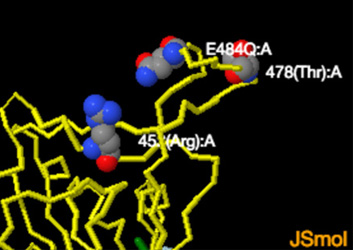
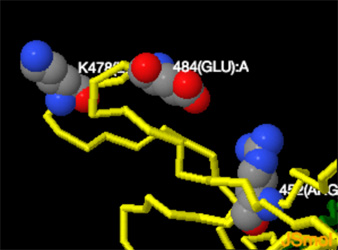
Note: Jsmol structures below -- mutations performed using Swiss-Pdb -- no further minimizations or Molecular Dynamics Simulations were performed
PDB:7v7f-->B.1.617.1 kappa variant showing Glyc(614); Thr(478); E484Q mutation; Arg(452)
-------------->spin on -------->- spin off
->zoom in to E484Q -------->- zoom out
To Rotate the Molecule--->Left Click and Drag
To Zoom-->>Left Click + hold Shift button and Drag Vertically
To see amino acid and atom number hold cursor over atom
Jmol Menu --->>Right-Click
PDB:77v7f-->mutated to B.1.617.2--> D614G, T478K, L452R, P681R. --E484Q--> E484(Glu) -
-------------->spin on -------->- spin off
->zoom in to E484 (Glu) -------->- zoom out
To Rotate the Molecule--->Left Click and Drag
To Zoom-->>Left Click + hold Shift button and Drag Vertically
To see amino acid and atom number hold cursor over atom
Jmol Menu --->>Right-Click
How Dangerous is the Delta Variant (B.1.617.2
The receptor binding domain is the portion of the spike protein that binds directly to human ACE2 receptors. Delta has 3 RBD mutations. The first, a lysine to asparagine substitution at position 417, is present in some, but not all sequences of B.1.617.2. It is also common to the Beta variant and has been associated with conformational changes to S protein, which may aid in immune escape. The second mutation, a leucine to arginine substitution at position 452, is common to the former variant of interest Epsilon, and is known to increase affinity for ACE2 receptors found on the surface of a variety of human cells, including the lungs. And the third, a threonine to lysine substitution at position 478, is common to the B.1.1.519 lineage, and has been predicted to increase electrostatic potential and steric hindrance, which may further increase RBD/ACE2 binding affinity and enable immune escape. ..."How Dangerous Is the Delta Variant (B.1.617.2)?
-------------->spin on -------->- spin off
-------------->zoom into receptor binding site
-(Amino acids 417K ,478T, 452L)
------->- zoom out
Jmol Menu --->>Right-Click
-------------->spin on -------->- spin off
-------------->zoom into receptor binding site
-(Amino acids 417N (this is delta+), 478K, 452R ---Delta with K417N originally corresponded to lineages AY.1 and AY.2
------->- zoom out
Jmol Menu --->>Right-Click
Readings:
Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses Nov. 2021
SARS-CoV-2 Variant Classifications and Definitions Oct. 2021
Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant --Oct 2021
Structural and functional mechanism of SARS-CoV-2 cell entry
Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant Aug. 2021
Sars-CoV-2 Variants
The novel coronavirus SARS-CoV2
B.1.1.7 and B.1.525 UK variant
B.1.427/B.1.429, California QP77P Mutation
B.1.526, NY E484K or S477N Mutations
Molecules of Disease
-
Small Molecules
Cholesterol
Nicotine
Trans Fatty Acid
Alcohol
Acetyaldehyde
- Proteins
Cytokines
Hemoglobin S
Prions
Botulinum Toxin
Explain it with Molecules
- Why is water such a good solvent?
- Why does ice float?
- Why do solids, liquids and gases behave differently?
- What is the geometry of methane?
- What's the difference between alpha and beta glucose?
- How does caffeine work in the brain?
- How does soap work?
- What is the difference between sucrose and fructose?
- Why is carbon monoxide so dangerous?
- Why is graphite so soft if it is made of only carbon?
- What is the difference between Carbyne and Graphite?
- Why is the fullerene and similar structures the cornerstone of nanotechnology?
- How big is a nanotube?
- Why does table salt have a cubic crystal shape?
- What is the structure of the benzene molecule?
- Why do carcinogens cause cancer?
- What causes Sickle Cell Anemia?
- What is the difference between sodium nitrite and nitrate?
- How do drugs work?